Background and aim:
Most conventional molecular diagnostics usually detect only a single disease-related biomarker. Great examples are the PCR tests currently used to diagnose COVID-19 by detecting a specific sequence from SARS-CoV-2.
Such so-called singleplex methods provide reliable results because they are “calibrated” to a single biomarker. However, determining whether a patient is infected with a new SARS-CoV-2 variant or a completely different pathogen requires probing for many different biomarkers at one time.
Scientists from the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius Maximilians University (JMU) in Würzburg have now developed a new diagnostic platform with LEOPARD. LEOPARD, which stands for “Leveraging Engineered tracrRNAs and On-target DNAs for PArallel RNA Detection,” is based on the finding that DNA cutting by Cas9 could be linked to the presence of a specific ribonucleic acid (RNA). This link allows LEOPARD to detect many RNAs at once, opening opportunities for the simultaneous detection of RNAs from viruses and other pathogens in a single patient sample.
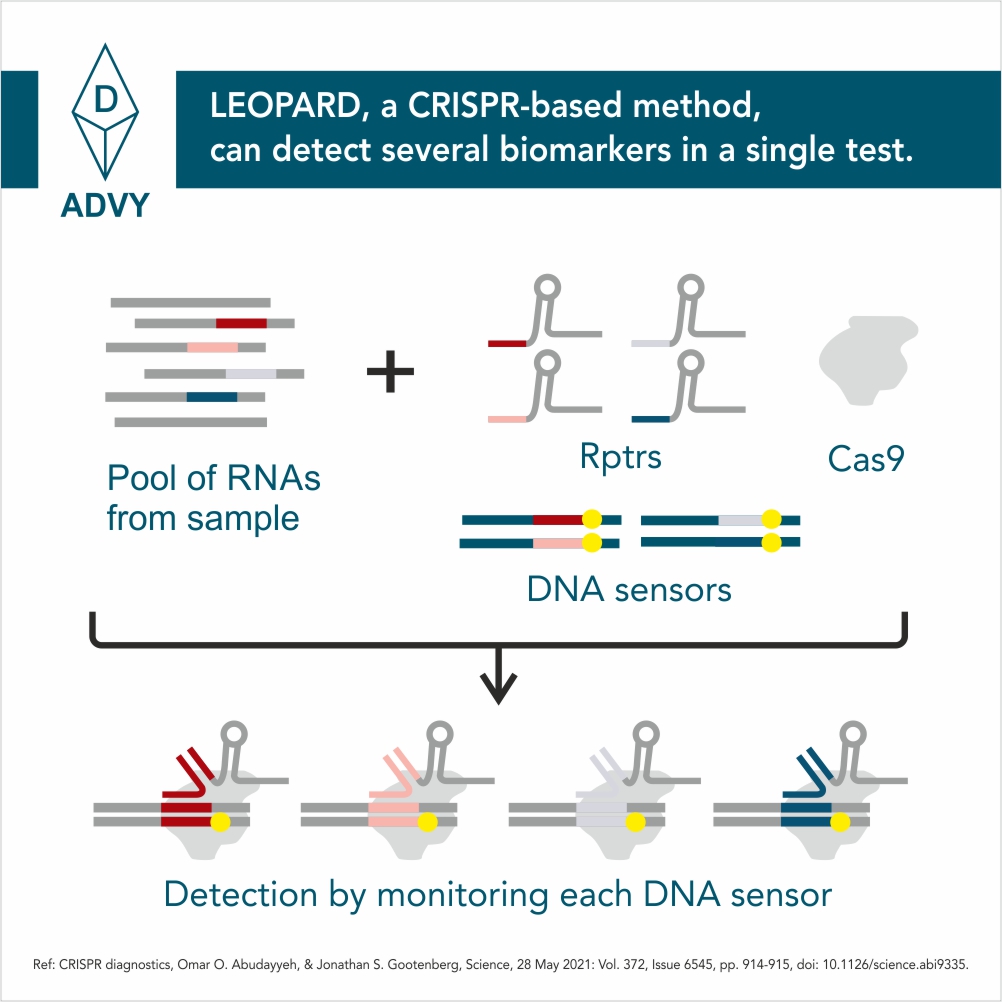
CRISPR-Cas9 is principally known as a bio-molecular tool for genome editing. Here, CRISPR-Cas9 functions as molecular scissors that cut specific DNA sequences. These same scissors are naturally used by bacteria to cut DNA associated with invading viruses.
Whether editing genomes or eliminating viruses, Cas9 cutting is directed by guide RNAs. The guide RNAs found in bacteria must pair with a separate RNA called the tracrRNA. The RNA couple then can work with Cas9 to direct DNA cutting.
Ref: News Medical